News

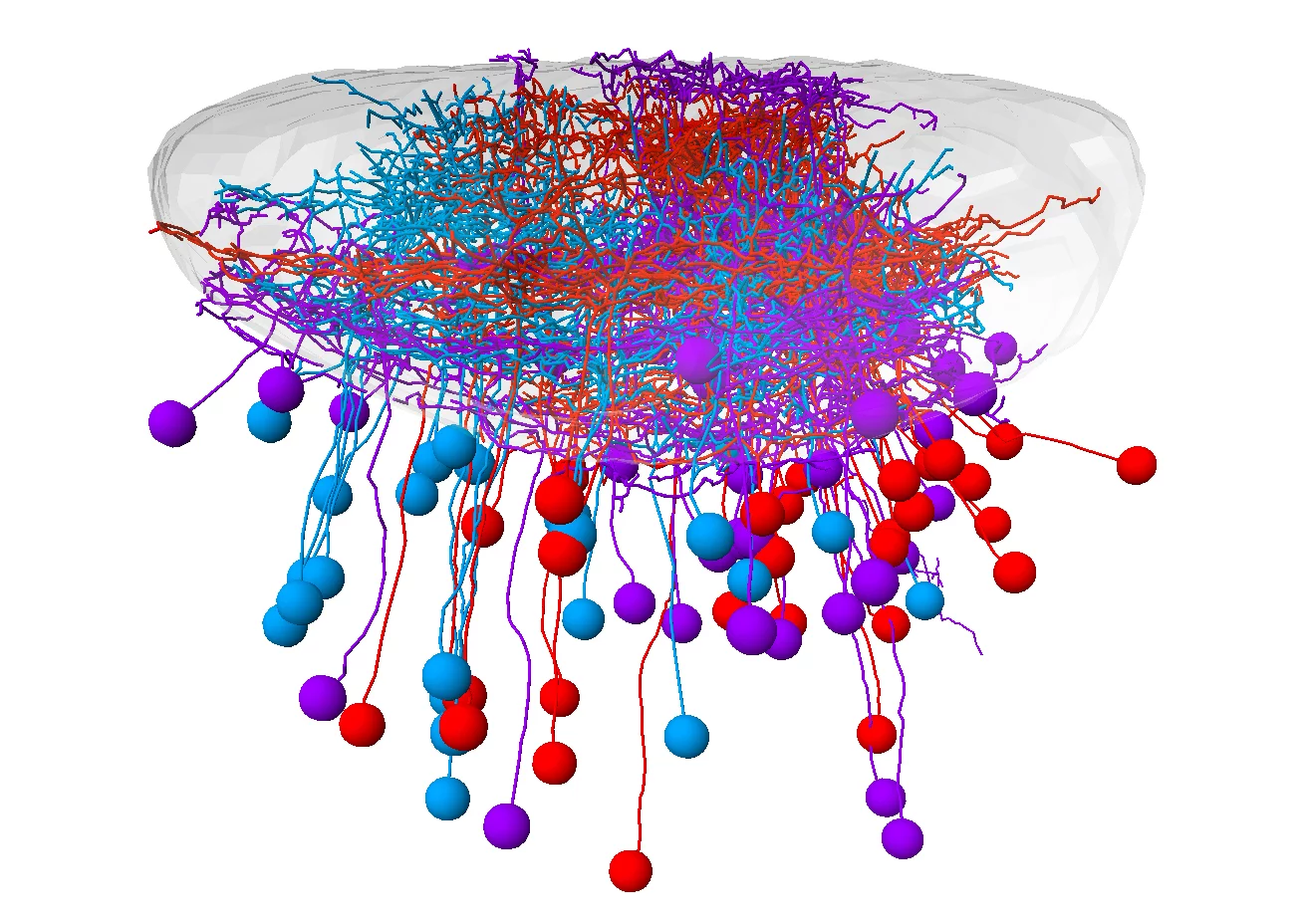
Retinal ganglion cells (RGCs) are the bottleneck through which all visual impressions flow from the retina to the brain. A team from the Max Planck Institute of Neurobiology, University of California Berkeley and Harvard University created a molecular catalog that describes the different types of these neurons. In this way, individual RGC types could be systematically studied and linked to a specific connection, function and behavioral response.
When zebrafish see light, they often swim towards it. Same with prey, although the signals are entirely different. A predator, on the other hand, prompts the fish to escape. That’s good, because a mix-up would have fatal consequences. But how does the brain manage to react to a visual stimulus with the proper behavior?
Optical signals are generated by photons that bombard the retina of the eye. Neurons in the retina collect and process these impressions. While doing so, the retina focuses on the important details: Is there contrast or color? Are there small or large objects? Is something moving? Once these details are filtered out, retinal ganglion cells (RGCs) send them to the brain, where they are translated into a specific behavior.
As the only connection between the retina and the brain, RGCs play a central role in the visual system. We already knew that specific RGC types sends different details to different regions of the brain. However, it has been unclear how RGC types differ on the molecular level, what their respective functions are, and how they help to regulate context-dependent behavior.

Klein, A.S., and Gogolla, N.
Science, 2021, 371, 122-123.
doi: 10.1126/science.abf5940
How mice feel each other's pain or fear
Empathic behaviors play crucial roles in human society by regulating social interactions, promoting cooperation toward a common goal, and providing the basis for moral decision-making (1, 2). Understanding the neural basis of empathy is crucial to understanding not only the human mind but also the neural mechanisms that give rise to social behaviors and the principles of our societies. Functional imaging studies in humans have identified essential brain regions that are engaged when people empathize with the affective experiences of others. However, human neuroimaging studies provide only limited spatial resolution and are solely correlative in nature. It has thus remained unclear how empathy with distinct affective experiences is set apart within the brain. read more

Hansen, F.M., Tanzer, M.C., Brüning, F., Bludau, I., Stafford, C., Schulman, B.A., Robles, M.S., Karayel, O., and Mann, M.
(IMPRS-LS students are in bold)
Nat Commun, 2021, 12, 254.
doi: 10.1038/s41467-020-20509-1
Data-independent acquisition method for ubiquitinome analysis reveals regulation of circadian biology
Protein ubiquitination is involved in virtually all cellular processes. Enrichment strategies employing antibodies targeting ubiquitin-derived diGly remnants combined with mass spectrometry (MS) have enabled investigations of ubiquitin signaling at a large scale. However, so far the power of data independent acquisition (DIA) with regards to sensitivity in single run analysis and data completeness have not yet been explored. Here, we develop a sensitive workflow combining diGly antibody-based enrichment and optimized Orbitrap-based DIA with comprehensive spectral libraries together containing more than 90,000 diGly peptides. This approach identifies 35,000 diGly peptides in single measurements of proteasome inhibitor-treated cells - double the number and quantitative accuracy of data dependent acquisition. Applied to TNF signaling, the workflow comprehensively captures known sites while adding many novel ones. An in-depth, systems-wide investigation of ubiquitination across the circadian cycle uncovers hundreds of cycling ubiquitination sites and dozens of cycling ubiquitin clusters within individual membrane protein receptors and transporters, highlighting new connections between metabolism and circadian regulation.

Brunner, A.-D., Thielert, M., Vasilopoulou, C., Ammar, C., Coscia, F., Mund, A., Horning, O.B., Bache, N., Apalategui, A., Lubeck, M., Raether, O., Park, M.A., Richter, S., Fischer, D.S., Theis, F.J., Meier, F., and Mann, M.
(IMPRS-LS students are in bold)
bioRxiv, 2020.2012.2022.423933.
doi: 10.1101/2020.12.22.423933
Ultra-high sensitivity mass spectrometry quantifies single-cell proteome changes upon perturbation
Single-cell technologies are revolutionizing biology but are today mainly limited to imaging and deep sequencing. However, proteins are the main drivers of cellular function and in-depth characterization of individual cells by mass spectrometry (MS)-based proteomics would thus be highly valuable and complementary. Chemical labeling-based single-cell approaches introduce hundreds of cells into the MS, but direct analysis of single cells has not yet reached the necessary sensitivity, robustness and quantitative accuracy to answer biological questions. Here, we develop a robust workflow combining miniaturized sample preparation, very-low flow-rate chromatography and a novel trapped ion mobility mass spectrometer, resulting in a more than ten-fold improved sensitivity. We accurately and robustly quantify proteomes and their changes in single, FACS-isolated cells. Arresting cells at defined stages of the cell cycle by drug treatment retrieves expected key regulators such as CDK2NA, E2 ubiquitin ligases such as UBE2S and highlights potential novel ones. Comparing the variability in more than 420 single-cell proteomes to transcriptome data revealed a stable core proteome despite perturbation. Our technology can readily be applied to ultra-high sensitivity analysis of tissue material, including post-translational modifications and to small molecule studies.

Where in a nerve cell is a certain receptor protein located? Without an answer to this question, it is difficult to draw firm conclusions about the function of this protein. Two scientists at the Max Planck Institute of Neurobiology developed a method in the fruit fly that marks receptor proteins in selected cells. In this way, they gained new insights into the neuronal mechanisms of motion vision. In addition, the research community receives an innovative tool to label proteins of all kinds.
One of the most fundamental questions in neurobiology is how sensory inputs are processed within the neuronal circuits of the brain. Thereby, it is not only important to understand which neurons are connected via synapses, but also how they communicate with each other. Receptors play a decisive role in this process.
These special proteins sit in the membrane envelope of neurons and specifically at synapses, where they receive incoming signals from other cells. Depending on receptor type and position, they determine how the cells react to incoming information: are they activated or inhibited, and how quickly does this happen? To understand a neural network in its entirety, it is therefore essential to study receptors and their distribution in neurons. However, this is not an easy task.
Some established methods provide little or no information about the distribution of proteins. Other techniques allow the labelling of receptors artificially introduced into cells, but not of naturally occurring ones. Therefore, the PhD students Sandra Fendl and Renee Vieira from Alexander Borst's department used the genetic resources available in the fruit fly Drosophila and developed a method to label proteins.
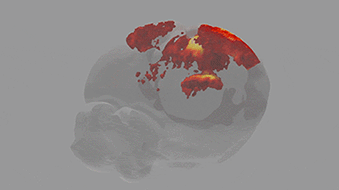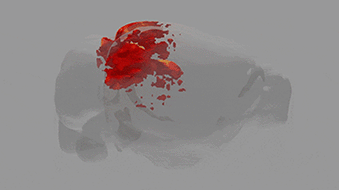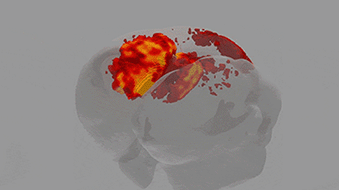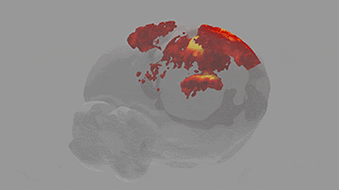
A team of scientists from the Neuro-Electronics Research Flanders developed and tested, in cooperation with scientists from the MPI of Neurobiology, a new volumetric functional ultrasound imaging platform. The ease of use, reliability, and affordability of the technology make it an excellent candidate for driving future brain-wide neuroimaging research.
Ultrasound is used to image soft tissue or organs, such as the heart, lungs and bladder, in real time. It is routinely used in hospitals because it is both safe and affordable. Over the past ten years, scientists at the Urban Lab (NERF, empowered by imec, KU Leuven and VIB) have contributed to the development of innovative brain ultrasound hardware and software solutions in collaboration with several academic and industrial partners. Using functional ultrasound imaging (fUSI) they have succeeded to visualize neural activity by mapping local changes in cerebral blood flow. Initially, however, fUSI was restricted to cross-sectional 2D imaging within a small field of view, which meant that visualization of brain-wide activity remained a challenge.
Spotting, pursuing and catching prey – for many animals this is an essential task for survival. Scientists at the Max Planck Institute of Neurobiology now show in zebrafish that the localization of neurons in the midbrain is adapted to a successful hunting sequence.
Far away, in the periphery of its visual field, a tiny zebrafish larva detects a small dot moving sideways. Is it prey or is it a threat, for instance, a distant predator sneaking up on it? Within the shortest possible time, the fish decides that it must be potential prey. The larva turns toward the object, approaches it, until it is right in front, and snaps shut – one of its daily hunting routines is successfully finished.
What might sound straightforward, is actually a highly complex process. Many different visual stimuli are detected simultaneously, transferred from the eye to the brain, and further processed. Interestingly, the stimuli don’t reach the brain at random locations: every position on the retina is transmitted to a very specific location in the tectum of the midbrain, the processing hub for visual stimuli. However, apart from that, there is not much knowledge of how the neurons are wired and organized, or which signals they specifically react to. Dominique Förster and a team from Herwig Baier’s laboratory analyzed how retinal ganglion cells transfer visual information from the eye to the tectum and how this input is further processed.

Weng, T.H., Steinchen, W., Beatrix, B., Berninghausen, O., Becker, T., Bange, G., Cheng, J., and Beckmann, R.
EMBO J, 2020, e105643, online ahead of print.
doi: 10.15252/embj.2020105643
Architecture of the active post-translational Sec translocon
In eukaryotes, most secretory and membrane proteins are targeted by an N-terminal signal sequence to the endoplasmic reticulum, where the trimeric Sec61 complex serves as protein-conducting channel (PCC). In the post-translational mode, fully synthesized proteins are recognized by a specialized channel additionally containing the Sec62, Sec63, Sec71, and Sec72 subunits. Recent structures of this Sec complex in the idle state revealed the overall architecture in a pre-opened state. Here, we present a cryo-EM structure of the yeast Sec complex bound to a substrate, and a crystal structure of the Sec62 cytosolic domain. The signal sequence is inserted into the lateral gate of Sec61α similar to previous structures, yet, with the gate adopting an even more open conformation. The signal sequence is flanked by two Sec62 transmembrane helices, the cytoplasmic N-terminal domain of Sec62 is more rigidly positioned, and the plug domain is relocated. We crystallized the Sec62 domain and mapped its interaction with the C-terminus of Sec63. Together, we obtained a near-complete and integrated model of the active Sec complex.

