News

Yildizoglu, T., Riegler, C., Fitzgerald, J.E., and Portugues, R.
Curr Biol, 2020, [Epub ahead of print].
doi: 10.1016/j.cub.2020.04.043
A Neural Representation of Naturalistic Motion-Guided Behavior in the Zebrafish Brain
All animals must transform ambiguous sensory data into successful behavior. This requires sensory representations that accurately reflect the statistics of natural stimuli and behavior. Multiple studies show that visual motion processing is tuned for accuracy under naturalistic conditions, but the sensorimotor circuits extracting these cues and implementing motion-guided behavior remain unclear. Here we show that the larval zebrafish retina extracts a diversity of naturalistic motion cues, and the retinorecipient pretectum organizes these cues around the elements of behavior. We find that higher-order motion stimuli, gliders, induce optomotor behavior matching expectations from natural scene analyses. We then image activity of retinal ganglion cell terminals and pretectal neurons. The retina exhibits direction-selective responses across glider stimuli, and anatomically clustered pretectal neurons respond with magnitudes matching behavior. Peripheral computations thus reflect natural input statistics, whereas central brain activity precisely codes information needed for behavior. This general principle could organize sensorimotor transformations across animal species.
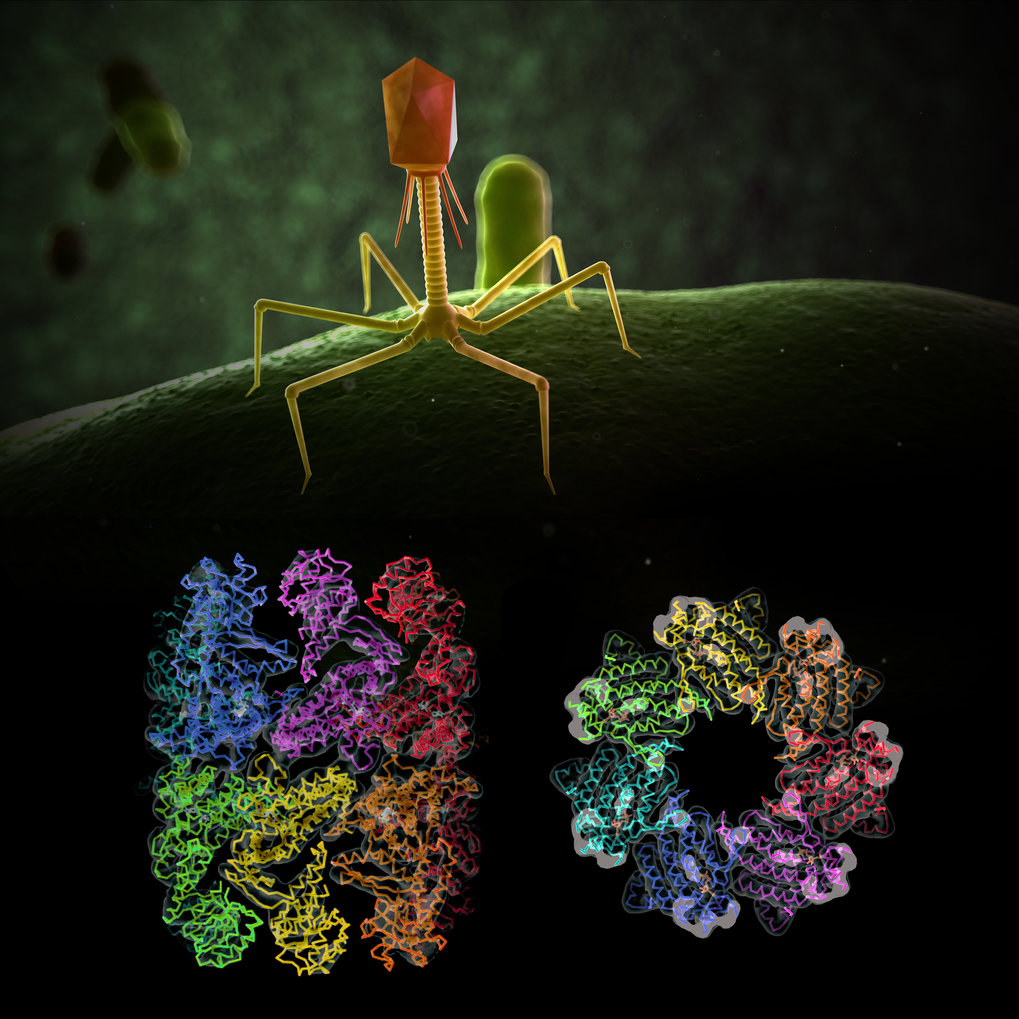
Researchers for protein folding helpers at the Max Planck Institute of Biochemistry have now deciphered the molecular structure of the EL phage chaperonin and discovered special features.
In general, viruses carry only a minimal amount of genetic information. They infect a host cell and use its metabolism and protein production machinery to reproduce. The bacteriophage EL also carries the genetic information of a protein folding helper, a so-called chaperonin. The Bacteriophage EL is a virus that can infect one of the most well-known hospital germs - the bacterium Pseudomonas aeruginosa. Researchers in the team of Manajit Hayer-Hartl and experts for protein folding helpers at the Max Planck Institute of Biochemistry have now deciphered the molecular structure of the EL phage chaperonin and discovered special features. The results of the crystal and cryo-electron microscopy structures show complexes with 7 or 14 subunits that form single and double rings. "In contrast to the known chaperonins, all the structures found were open, which means that they do not form a folding cage as in GroEL/GroES," says Andreas Bracher, first author of the study. "It is possible that this phage chaperonin represents a primitive, evolutionary precursor of today's cellular chaperonins, which works without encapsulation of the substrate proteins". The study was published in the science journal PLOS ONE.

Bittmann, J., Grigaitis, R., Galanti, L., Amarell, S., Wilfling, F., Matos, J., and Pfander, B.
(IMPRS-LS students are in bold)
Elife, 2020, 9.
doi: 10.7554/eLife.52459
An advanced cell cycle tag toolbox reveals principles underlying temporal control of structure-selective nucleases
Cell cycle tags allow to restrict target protein expression to specific cell cycle phases. Here, we present an advanced toolbox of cell cycle tag constructs in budding yeast with defined and compatible peak expression that allow comparison of protein functionality at different cell cycle phases. We apply this technology to the question of how and when Mus81-Mms4 and Yen1 nucleases act on DNA replication or recombination structures. Restriction of Mus81-Mms4 to M phase but not S phase allows a wildtype response to various forms of replication perturbation and DNA damage in S phase, suggesting it acts as a post-replicative resolvase. Moreover, we use cell cycle tags to reinstall cell cycle control to a deregulated version of Yen1, showing that its premature activation interferes with the response to perturbed replication. Curbing resolvase activity and establishing a hierarchy of resolution mechanisms are therefore the principal reasons underlying resolvase cell cycle regulation.

Max Planck researchers have for the first time developed a genome the size of a minimal cell that can copy itself.
The field of synthetic biology does not only observe and describe processes of life but also mimics them. A key characteristic of life is the ability to ability for replication, which means the maintenance of a chemical system. Scientists at the Max Planck Institute of Biochemistry in Martinsried generated a system, which is able to regenerate parts of its own DNA and protein building blocks. The results have now been published in Nature Communications.
In the field of synthetic biology, researchers investigate so-called “bottom-up” processes, which means the generation of life mimicking systems from inanimate building blocks. One of the most fundamental characteristics of all living organism is the ability to conserve and reproduce itself as distinct entities. However, the artificial “bottom-up” approach to create a system, which is able to replicate itself, is a great experimental challenge. For the first time, scientists have succeeded in overcoming this hurdle and synthesizing such a system.
A biological machine produces its own building blocks
Hannes Mutschler, head of the research group "Biomimetic Systems" at the MPI for Biochemistry, and his team are dedicated to imitate the replication of genomes and protein synthesis with a “bottom-up” approach. Both processes are fundamental for the self-preservation and reproduction of biological systems. The researchers now succeeded in producing an in vitro system, in which both processes could take place simultaneously. "Our system is able to regenerate a significant proportion of its molecular components itself," explains Mutschler. In order to start this process, the researchers needed a construction manual as well as various molecular "machines" and nutrients. Translated into biological terms, this means the construction manual is DNA, which contains the information to produce proteins. Proteins are often referred to as "molecular machines" because they often act as catalysts, which accelerate biochemical reactions in organisms.

Blessing, C., and Ladurner, A.G.
Nat Struct Mol Biol, [Epub ahead of print].
doi: 10.1038/s41594-020-0412-x
Tickling PARPs into serine action
Poly-(ADP-ribosylation) is a post-translational modification with broad roles in cell signaling. A recently reported crystal structure reveals how the accessory factor HPF1 extends the catalytic active site of PARP1 and PARP2 to promote the specific ADP-ribosylation of serine residues, a prerequisite for dynamic chromatin changes induced by DNA damage.

Rüdiger Klein receives an ERC Advanced Grant to analyze the development of amygdala circuits
The amygdala is important for the emotional evaluation of situations or objects. Aided by the amygdala, we are able learn to treat something with affection or aversion. How the amygdala controls the various behavioral reactions and which other brain regions are involved, remains unclear. In order to fill this gap in our knowledge, Rüdiger Klein attempts to specifically reorganize the development of the amygdala circuits in mice, thereby transforming innate and learned emotional behavior. The European Research Council (ERC) is funding this innovative project with a 2.5 million Euro grant over the next five years. Fear protects us from danger, but can also stand in our way when it becomes irrational. A good feeling from eating encourages us to choose suitable food, but can also contribute to eating disorders. No matter whether it is fear, pleasure or other emotions, neurons in the amygdala link our feelings with internal and external stimuli and thereby control our unconscious behavior.

Kober-Hasslacher, M., Oh-Strauß, H., Kumar, D., Soberón, V., Diehl, C., Lech, M., Engleitner, T., Katab, E., Fernandez Saiz, V., Piontek, G., Li, H., Menze, B., Ziegenhain, C., Enard, W., Rad, R., Böttcher, J.P., Anders, H.J., Rudelius, M., Schmidt-Supprian, M. (IMPRS-LS students are in bold)
J Clin Invest, 2020, [Epub ahead of print].
DOI: 10.1172/JCI124382
c-Rel gain in B cells drives germinal center reactions and autoantibody production
Single nucleotide polymorphisms and locus amplification link the NF-κB transcription factor c-Rel to human autoimmune diseases and B cell lymphomas, respectively. However, the functional consequences of enhanced c-Rel levels remain enigmatic. Here, we overexpressed c-Rel specifically in mouse B cells from BAC-transgenic gene loci and demonstrate that c-Rel protein levels linearly dictated expansion of germinal center (GC) B cells and isotype-switched plasma cells. c-Rel expression in B cells of otherwise c-Rel-deficient mice fully rescued terminal B cell differentiation, underscoring its critical B cell-intrinsic roles. Unexpectedly, in GCB cells transcription-independent regulation produced the highest c-Rel protein levels amongst B cell subsets. In c-Rel overexpressing GCB cells this caused enhanced nuclear translocation, a profoundly altered transcriptional program and increased proliferation. Finally, we provide a link between c-Rel gain and autoimmunity by showing that c-Rel overexpression in B cells caused autoantibody production and renal immune complex deposition.

Libicher, K., Hornberger, R., Heymann, M., and Mutschler, H.
Nat Commun, 2020, 11, 904.
doi: 10.1038/s41467-020-14694-2
In vitro self-replication and multicistronic expression of large synthetic genomes
The generation of a chemical system capable of replication and evolution is a key objective of synthetic biology. This could be achieved by in vitro reconstitution of a minimal self-sustaining central dogma consisting of DNA replication, transcription and translation. Here, we present an in vitro translation system, which enables self-encoded replication and expression of large DNA genomes under well-defined, cell-free conditions. In particular, we demonstrate self-replication of a multipartite genome of more than 116 kb encompassing the full set of Escherichia coli translation factors, all three ribosomal RNAs, an energy regeneration system, as well as RNA and DNA polymerases. Parallel to DNA replication, our system enables synthesis of at least 30 encoded translation factors, half of which are expressed in amounts equal to or greater than their respective input levels. Our optimized cell-free expression platform could provide a chassis for the generation of a partially self-replicating in vitro translation system.

